Abstract
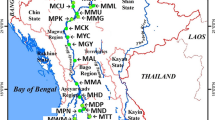
Pakistan has natural freshwater resources acting as a hotspot for diverse fish fauna. However, this aquatic fauna is declining at an alarming rate due to over-exploitation, habitat degradation, water pollution, climate change, and certain anthropogenic activities. The freshwater shark, Wallago attu, is a popular edible catfish inhabiting these freshwater ecosystems. Habitat degradation, overfishing, and human activities are heavily impacting the natural population of this species. So, sound knowledge about its population structure is necessary for its proper management in natural waters. The current study involves utilizing two mtDNA markers (COI, Cytb) to assess the genetic structure and differentiation among W. attu populations of Pakistani Rivers. Genetic variability analysis indicated a high haplotype (0.343 ± 0.046–0.870 ± 0.023) and low nucleotide diversity (0.0024 ± 0.012–0.0038 ± 0.018) among single and combined gene sequences, respectively. Overall, River Indus was populated with more diverse fauna of Wallago attu as compared to River Chenab and River Ravi. Population pairwise, Fst values (0.40–0.61) were found to be significantly different (p < 0.01) among three Riverine populations based upon combined gene sequences. The gene flow for the combined gene (COI + Cytb) dataset among three populations was less than 1.0. The transition/transversion bias value R (0.58) was calculated for testing of neutral evolution, and it declared low genetic polymorphism among natural riverine populations of Wallago attu. The current study’s findings would be meaningful in planning the management and conservation of this economically important catfish in future.






Similar content being viewed by others
Data availability
All data generated or analyzed during the study are included in the manuscript.
References
Akib NAM, Tam BM, Phumee P, Abidin MZ, Tamadoni S, Mather PB, Nor SAM (2015) High connectivity in Rastrelliger kanagurta: influence of historical signatures and migratory behaviour inferred from mtDNA cytochrome b. PLoS One 10(3):e0119749. https://doi.org/10.1371/journal.pone.0119749
Barrett RDH, Schluter D (2008) Adaptation from standing genetic variation. Trends Ecol Evol 23(1):38–44. https://doi.org/10.1016/j.tree.2007.09.008
Basharat H, Ghafoor A, Chavhan A, Zafar M, Abbas K, Parveen J (2016) Microsatellite markers revealed poor genetic structure of Wallago attu in Punjab, Pakistan. Int J Life Sci 4(3):385–393
Behera BK, Baisvar VS, Rout AK, Pakrashi S, Kumari K, Panda D, Das P, Parida PK, Meena DK, Bhakta D, Das BK, Jena JK (2017) The population structure and genetic divergence of Labeo gonius (Hamilton, 1822) analyzed through mitochondrial DNA cytochrome b gene for conservation in Indian waters. Mitochondrial DNA Part A 29(4):543–551. https://doi.org/10.1080/24701394.2017.1320992
Behera BK, Kunal SP, Baisvar VS, Meena DK, Panda D, Pakrashi S, Paria P, Das P, Debnath D, Parida PK, Das BK, Jena JK (2018) Genetic variation in wild and hatchery population of Catla catla (Hamilton, 1822) analyzed through mtDNA cytochrome b region. Mitochondrial DNA Part A 29(1):126–131. https://doi.org/10.1080/24701394.2016.1253072
Beheregaray L, Attard C, Brauer C, Hammer M (2016) Innovations in conservation: How genetics can help save freshwater fishes. Wildl Aust 53(3):34–37
Biswas I, Nagesh TS, Sajina AM (2019) Stock delineation in Clupisoma garua (Hamilton, 1822) populations of Ganga Riverine system using truss network analysis. Indian J Fish 66(2):1–7
Carvalho G, Hauser L (1994) Molecular genetics and the stock concept in fisheries. Rev Fish Biol Fish 4:326–335
Chanthran SSD, Lim PE, Li Y, Liao TY, Poong SW, Du J, Hussein MAS, Sade A, Rumpet R, Loh KH (2020) Genetic diversity and population structure of Terapon jarbua (Forskal, 1775)(Teleostei, Terapontidae) in Malaysian waters. ZooKeys 911:139
Das SP, Swain S, Jena J, Das P (2018) Genetic diversity and population structure of Cirrhinus mrigala revealed by mitochondrial ATPase 6 gene. Mitochondrial DNA Part A 29(4):495–500
Excoffier L, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinf 1:117693430500100000
Excoffier L, Smouse PE, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131(2):479–491
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587. https://doi.org/10.1093/genetics/164.4.1567
Fang DA, Huiluo MH, Mao C, Kuang Z, Qi H, Xu D, Tan L, Li Y (2023) Genetic diversity and population differentiation of naked carp (Gymnocypris przewalskii) revealed by cytochrome oxidase subunit I and d-loop. Front Ecol Evol 11(10):245
Frankham R, Brook BW, Bradshaw CJA, Traill LW, Spielman D (2013) 50/500 rule and minimum viable populations: response to Jamieson and Allendorf. Trends Ecol Evol 28:187–188
Funk WC, Forester BR, Converse SJ, Darst C, Morey S (2019) Improving conservation policy with genomics: a guide to integrating adaptive potential into U.S. Endangered Species Act decisions for conservation practitioners and geneticists. Conserv Genet 20:115–134. https://doi.org/10.1007/s10592-018-1096-1
Gupta A, Lal KK, Mohindra V, Singh RK, Punia P, Dwivedi AK, Gupta BK, Luhariya RK, Masih P, Mishra RM, Jena JK (2013) Genetic divergence in natural populations of bronze featherback, Notopterus notopterus (Osteoglossiformes: Notopteridae) from five Indian Rivers, analyzed through mtDNA ATPase6/8 regions. Meta Gene 1:50–57
Habib M, Lakra WS, Mohindra V, Lal KK, Punia P, Singh RK, Khan AA (2012) Assessment of ATPase 8 and ATPase 6 mtDNA sequences in genetic diversity studies of Channa marulius (Channidae: Perciformes). Proc Natl Acad Sci B Biol Sci 82:497–501
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hogan ZS (2011) Ecology and conservation of large-bodied freshwater catfish: a global perspective. Am Fish Soc Symp 77:39–53
Hudson AG, Vonlanthen P, Seehausen O (2014) Population structure, inbreeding and local adaptation within an endangered riverine specialist: the nase (Chondrostoma nasus). Conserv Genet 15:933–951
Iqbal MM, Shoaib M, Agwanda P, Lee JL (2018) Modeling approach for water-quality management to control pollution concentration: a case study of Ravi River, Punjab, Pakistan. Water 10(8):1068
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Kumari P, Pavan-Kumar A, Kumar G, Alam A, Parhi J, Gireesh-Babu P, Chaudhari A, Krishna G (2017) Genetic diversity and demographic history of the giant River catfish Sperata seenghala inferred from mitochondrial DNA markers. Mitochondrial DNA Part A 28(6):920–926
Li XY, Deng YP, Yang K, Gan WX, Zeng RK, Deng LJ, Song ZB (2016) Genetic diversity and structure analysis of Percocypris pingi (Cypriniformes: Cyprinidae): implications for conservation and hatchery release in the Yalong River. PLoS One 11(12):e0166769
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11):1451–1452
Liu BH (2017) Statistical genomics: linkage, mapping, and QTL analysis. CRC Press. ISBN: 9780367400743. https://doi.org/10.1201/9780203738658
Loreille O, Roumat E, Verneau O, Bouchet F, Hänni C (2001) Ancient DNA from Ascaris: extraction amplification and sequences from eggs collected in coprolites. Int J Parasitol 31(10):1101–1106
Mirza MR (2003) Checklist of freshwater fishes of Pakistan. Pak J Zool Suppl Series 3:1–30
Mirza MR, Sandhu AA (2007) Fishes of the Punjab Pakistan. Polymer Publication, Lahore, Pakistan
Montana CG, Choudhary SK, Dey S, Winemiller KO (2011) Compositional trends of fisheries in the River Ganges, India. Fish Manag Ecol 18:282–296
Navarro-Ortega A, Acuna V, Bellin A, Burek P, Cassiani G, Choukr-Allah R, Barcelo D (2015) Managing the effects of multiple stressors on aquatic ecosystems under water scarcity. The GLOBAQUA project. Sci Total Environ 503:3–9
Ng HH, Alwis Goonatilake S, Fernado M, Kotagama O (2019) Wallago attu (errata version published in 2020). The IUCN Red List of Threatened Species, e.T166468A174784999
Oleksiak MF (2010) Genomic approaches with natural fish populations. J Fish Biol 76:1067–1093
Ozcelik H, Shi X, Chang MC, Tram E, Vlasschaert M, Di Nicola N, Siminovitch K (2012) Long-range PCR and next-generation sequencing of BRCA1 and BRCA2 in breast cancer. J Mol Diagn 14(5):467–475
Porrini LP, Iriarte PJF, Iudica CM, Abud EA (2015) Population genetic structure and body shape assessment of Pagrus pagrus (Linnaeus, 1758) (Perciformes: Sparidae) from the Buenos Aires coast of the Argentine Sea. Neotrop Ichthyol 13(2):431–438
Ramos-Onsins SE, Rozas J (2002) Statistical properties of new neutrality tests against population growth. Mol Biol Evol 19:2092–2100
Renjithkumar CR, Roshni K, Kurup BM (2016) Exploited fishery resources of Muvattupuzha River, Kerala, India. Fish Tech 53:177–182
Sahoo L, Mohanty M, Meher PK, Murmu K, Sundaray JK, Das P (2019) Population structure and genetic diversity of hatchery stocks as revealed by combined mtDNA fragment sequences in Indian major carp, Catla catla. Mitochondrial DNA Part A 30(2):289–295
Sala OE, Chapin FS, Armesto JJ, Berlow E, Bloomfield J, Dirzo R, HuberSanwald E, Huenneke LF, Jackson RB, Kinzig A, Leemans R (2000) Biodiversity global biodiversity scenarios for the year 2100. Science 287(5459):1770–1774
Singh RK, Lal KK, Mohindra V, Punia P, Sah RS, Kumar R, Gupta A, Das R, Lakra WS, Ayyappan S (2012) Genetic diversity of Indian Major Carp, Labeo calbasu (Hamilton, 1822) populations inferred from microsatellite loci. Biochem Syst Ecol 44:307–316
Song Z, Song J, Yue B (2008) Population genetic diversity of Prenant’s schizothoracin, Schizothorax prenanti, inferred from the mitochondrial DNA control region. Environ Biol Fish 81:247–252
Srihari M, Kathirvelpandian A, GiriBhavan S, Sajina AM, Gangan SS (2019) Deciphering the stock structure of Chanos chanos (Forsskal, 1775) in Indian waters by truss network and otolith shape analysis. Turk J Fish Aquat Sci 20(2):103–111
Vieira AR, Rodrigues ASB, Sequeira V, Neves A, Paiva RB, Paulo OS, Gordo LS (2016) Genetic and morphological variation of the forkbeard, Phycis phycis (Pisces, Phycidae): evidence of panmixia and recent population expansion along its distribution area. PLoS One 11(12):e0167045
Vila M, Hermida M, Fernan C, Perea S, Doadrio I, Amaro R, San ME (2017) Phylogeography and conservation genetics of the ibero-Balearic threespined stickleback (Gasterosteus aculeatus). PLoS One 12(1):e0170685
Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PD (2005) DNA barcoding Australia’s fish species. Philos Trans R Soc Lond B Biol Sci 360(1462):1847–1857
Wright S (1965) The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 19:395–420
WWF Living Planet Report (2016) Risk and resilience in a new era. WWF International, Gland, Switzerland
Xiao W, Zhang Y, Liu H (2001) Molecular systematics of Xenocyprinae (Teleostei: Cyprinidae): taxonomy, biogeography, and coevolution of a special group restricted in East Asia. Mol Phylogenet Evol 18(2):163–173
Yousaf M, Salam A, Naeem M (2011) Body composition of freshwater Wallago attu in relation to body size, condition factor and sex from southern Punjab, Pakistan. Afr J of Biotech 10(20):4265–4268
Zhang X, Gao X, Wang JW, Cao WX (2015) Extinction risk and conservation priority analyses for 64 endemic fishes in the upper Yangtze River, China. Environ Biol Fish 98(1):261–272
Zhang C, Li Q, Wu X, Liu Q, Cheng Y (2018) Genetic diversity and genetic structure of farmed and wild Chinese mitten crab (Eriocheir sinensis) populations from three major basins by mitochondrial DNA COI and Cyt b gene sequences. Mitochondrial DNA Part A 29(7):1081–1089
Author information
Authors and Affiliations
Contributions
SS wrote the main manuscript. SS and SAK collected samples. SS and NH completed lab work. MAME performed data analysis, and AH reviewed the final draft of the manuscript. All authors have thoroughly read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethical approval
It has been assured that the experimental samples of fish species and the collection of fish species were carried out with relevant institutional, national, and international guidelines and legislation with appropriate permissions from competent authorities.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Responsible Editor: Bruno Nunes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sherzada, S., Hussain, N., Hussain, A. et al. Diversity and genetic structure of freshwater shark Wallago attu: an emerging species of commercial interest. Environ Sci Pollut Res 31, 15571–15579 (2024). https://doi.org/10.1007/s11356-024-32117-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-024-32117-3